Environmental DNA metabarcoding as an alternative to macrobenthic assessments in fish-farm compliance assessment
Salmon farming is a major industry in Scotland. With a turnover of in excess on £1b, it supports over 2500 direct jobs and around 10,000 total supply chain jobs in some of Scotland's most isolated and fragile rural areas, including the Highlands and Islands.
Salmon farming, like most farming, changes the receiving environment in several ways. Changes around salmon farms include those occurring on the seabed. To ensure regulatory compliance, fish-farmers are required to undertake seabed (benthic) monitoring around their sites. Until recently they had to monitor the seabed by taking seven grab-samples along a single transect per production cycle. Under a new regulatory framework, fish-farmers now have to collect and analyse 28 benthic samples across multiple transects. From each sample, macrobenthic taxonomic assessments are made and used to generate the 'infaunal quality index' (IQI) from which the benthic status and compliance is assessed. The four-fold increase in sampling effort (from 7 to 28 samples) is very expensive to deliver and is beyond the capacity of current service providers. In addition, macrobenthic faunal analysis is time-consuming (3 – 6 months) and the time-gap between sampling and data interpretation prevents active, near-real-time management of farm sites.
Metabarcoding is the process of identifying organisms via their unique DNA sequences, present in the environment (eDNA), and can be applied to sediment samples. Metabarcoding involves four steps: 1. eDNA extraction, 2. eDNA amplification of specific markers, 3. eDNA sequencing followed by 4. sequence annotation.
In previous, SAIC and SSPO supported, proof-of-concept research we subjected grab samples taken around fish-cages to both standard taxonomic- and metabarcoding (eDNA)-based analysis. We demonstrated that bacterial-based eDNA characterisation was precise and, using Random forest (RF) machine learning, that patterns of bacterial eDNA could be used to predict macrobenthic IQI.
The BactMetBar partnership of academic institutions, fishfarming companies and the regulator is innovating to deliver the following key objectives:
1. Refine, standardise and publish standard protocols in relation to eDNA extraction and amplification for Scottish benthic samples taken for eDNA-based analysis over a diverse range of habitats
2. Develop data-processing and RF algorithms that best predict the IQI from bacterial eDNA sequence data by accounting for spatial and temporal variability in the bacterial response to fish-farm associated change
3. Collate the statistical routines into a single, user-friendly ‘R-package’
4. Publicise the standard operating procedures (SOPs) and demonstrate the BactMetBar R-package and embed the innovative alternative approach to macrobenthic-based monitoring in the sector (knowledge exchange).
This project and its outputs have the potential to create a shift change in how the Scottish salmon farmers assess the impacts of their farms.
The benefits stretch beyond regulatory compliance assessment, supporting improved production efficiencies and the development of new farms through improved modelling.
Role of SAMS in the BactMetBar project
SAMS coordinates the project, working with the UHI Rivers and Lochs Institute to focus the sampling effort.
SAMS is bringing the data together, and trains the machine.
SAMS also develops and tests ways to optimise the machine's ability to correctly predict environmental status from eDNA.
SAMS and the BMB team have completed the core deliverable, the eDNA2IQI pipeline which is published here:
Wyness, A. J., Stoeck, T., Morrissey, B. J., Berrill, I., Davidson, K., FairBairn, C., MacIntyre, S., Webb, C., Foster, T., Brain, S., Svobodova, D., Clark, C., Ashley-Wheeler, V., Roosenmaallen, E., Pollard, P., & Wilding, T. A. (2026). eDNA2IQI: an eDNA/random forest-based prediction tool that accurately predicts ecological status for regulatory-compliance assessment. Aquaculture, 612. https://doi.org/10.1016/j.aquaculture.2025.743126
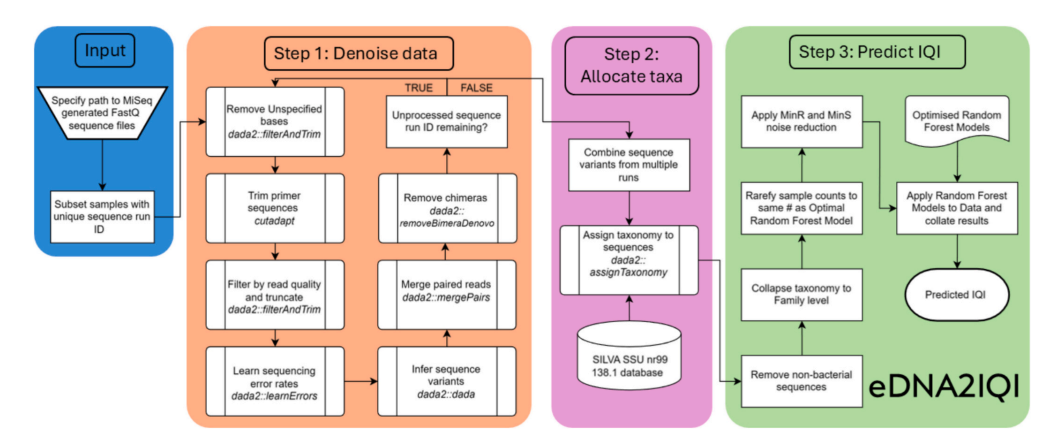
Flow diagram describing eDNA2IQI process for predicting IQI values from raw eDNA sequence data. Chart symbols follow American National Standards Institute (ANSI)/International Organization for Standardization (ISO) standards.
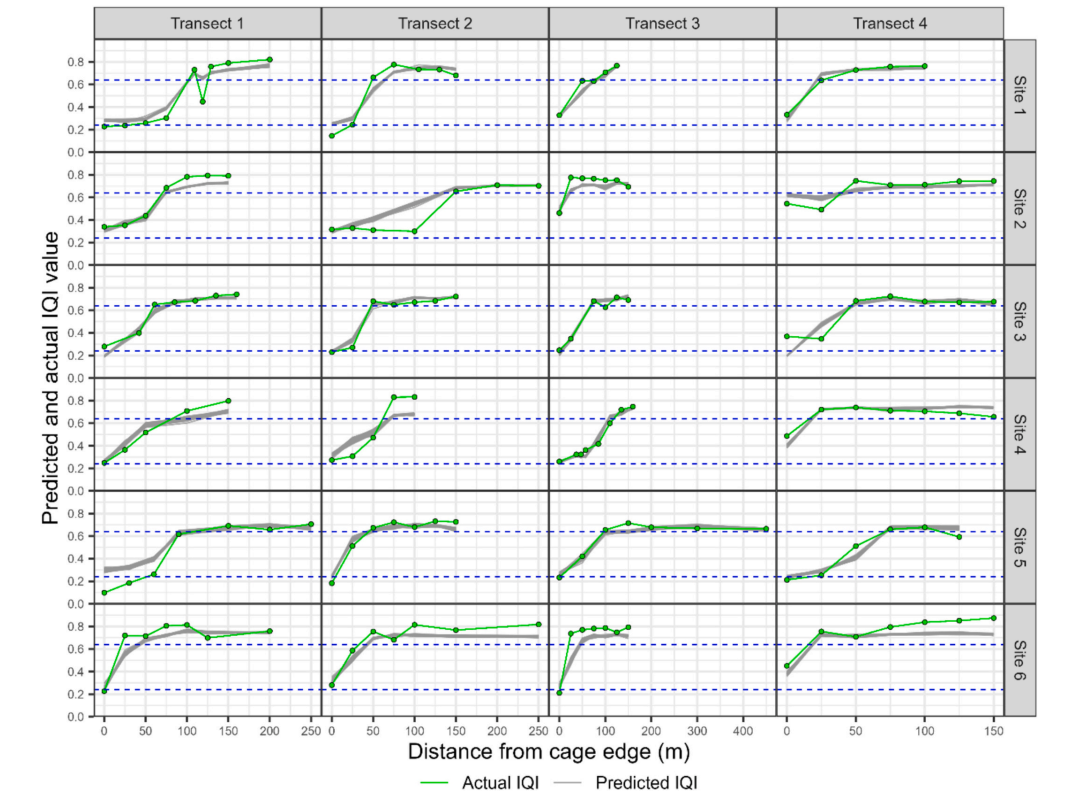
Six full site compliance survey exemplars using eDNA2IQI. The macrobenthic (actual) IQI is shown as a green line, while outputs from 30 RF models indicated as separate (overlapping) grey lines (varying thicknesses). Regulatory boundaries at 0.64 (moderate/good) and 0.24 (bad/moderate) are included for reference. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We have also demonstrated a high degree of congruity between different labs processing the same samples. This gives us confidence that the eDNA2IQI model is robust. Publication pending.
Relevant publications by the BactMetBar team
Wilding, T. A., T. Stoeck, B. J. Morrissey, S. F. Carvalho and M. W. Coulson (2023). "Maximising signal-to-noise ratios in environmental DNA-based monitoring." Science of The Total Environment 858.
Dully, V., G. Rech, T. A. Wilding, A. Lanzen, K. MacKichan, I. Berrill and T. Stoeck (2021). "Comparing sediment preservation methods for genomic biomonitoring of coastal marine ecosystems." Mar Pollut Bull 173(Pt B): 113129.
Dully, V., T. A. Wilding, T. Mühlhaus and T. Stoeck (2021). "Identifying the minimum amplicon sequence depth to adequately predict classes in eDNA-based marine biomonitoring using supervised machine learning." Computational and Structural Biotechnology Journal 19: 2256-2268.
Dully, V., H. Balliet, L. Frühe, M. Däumer, A. Thielen, S. Gallie, I. Berrill and T. Stoeck (2020). "Robustness, sensitivity and reproducibility of eDNA metabarcoding as an environmental biomonitoring tool in coastal salmon aquaculture – An inter-laboratory study." Ecological Indicators.
Frühe, L., T. Cordier, V. Dully, H.-W. Breiner, G. Lentendu, J. Pawlowski, C. Martins, T. A. Wilding and T. Stoeck (2020). "Supervised machine learning is superior to indicator value inference in monitoring the environmental impacts of salmon aquaculture using eDNA metabarcodes." Molecular Ecology DOI: 10.1111/mec.15434
Forster, D., G. Lentendu, S. Filker, E. Dubois, T. A. Wilding and T. Stoeck (2019). "Improving eDNA-based protist diversity assessments using networks of amplicon sequence variants." Environ Microbiol 21(11): 4109-4124.
Frühe, L., V. Dully, D. Forster, N. B. Keeley, O. Laroche, X. Pochon, S. Robinson, T. A. Wilding and T. Stoeck (2021). "Global Trends of Benthic Bacterial Diversity and Community Composition Along Organic Enrichment Gradients of Salmon Farms." Frontiers in Microbiology12(853).
Cordier, T., D. Forster, Y. Dufresne, C. I. M. Martins, T. Stoeck and J. Pawlowski (2018). "Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring." Mol Ecol Resour 18(6): 1381-1391
Lejzerowicz, F., P. Esling, L. Pillet, T. A. Wilding, K. D. Black and J. Pawlowski (2015). "High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems." Sci Rep 5: 13932.
Pawlowski, J., P. Esling, F. Lejzerowicz, T. Cedhagen and T. A. Wilding (2014). "Environmental monitoring through protist next-generation sequencing metabarcoding: assessing the impact of fish farming on benthic foraminifera communities." Molecular Ecology Resources14(6): 1129 - 1140.
Cordier, T., D. Forster, Y. Dufresne, C. I. M. Martins, T. Stoeck and J. Pawlowski (2018). "Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring." Mol Ecol Resour 18(6): 1381-1391.


